Pseudouridylation, uridine (U) isomerization to pseudoUridine (ψ), is the most common post-transcriptional modification of RNA nucleosides used to control gene expression in various cellular processes in living organisms. Therefore, ψ is considered to be the fifth ribonucleoside. ψ is expressed at high levels in both eukaryotic tRNA and rRNA.
PseudoUridine modifications contribute to:
- Stabilization of RNA structure and ribosome biogenesis and activity.
- Regulation of rRNA processing, pre-mRNA splicing and protein synthesis.
- Control of growth, development and response to stress in different organisms.
PseudoUridine sequencing methods
- Localisation in RNA analyzed by chromatography and mass spectrometry quantification.
- Revealing sites of pseudoUridine in the transcriptome by high-throughput NGS of pseudoUridine-specific chemistry.
- Oxford Nanopore Technology (ONT) or Pacific Biosciences' Third Generation Sequencing (TGS) technology sequences a single molecule of DNA or RNA to omit the introduction of bias in workflows such as selective chemistry, and reverse transcription and/or PCR, enabling direct observation of modified bases. (Fig. 1).
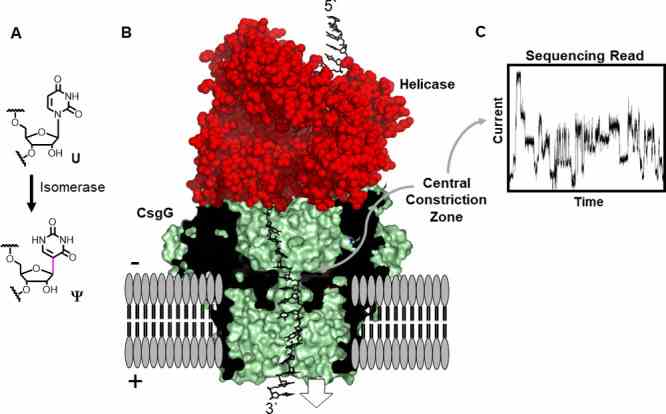 Fig. 1 Direct RNA sequencing for ψ by monitoring current vs time traces in a protein nanopore-helicase platform. (Fleming AM, 2021)
Fig. 1 Direct RNA sequencing for ψ by monitoring current vs time traces in a protein nanopore-helicase platform. (Fleming AM, 2021)
Sequencing and conformational assignment of pseudoUridine in SARS-CoV-2
Professor Cynthia J. Burrows' team at the Department of Chemistry, University of Utah, USA, has achieved pseudoUridine detection on sequence-unbiased RNA using a nanopore deconvolution enzyme system combined with blocking current and dwell time analysis methods.
The authors found that ψ affects the read residence time via the exonuclease active site compared to U. The residence time of U conforms to a Gaussian distribution, while ψ satisfies a double Gaussian distribution (Psi-1 and Psi-2). This finding revealed that the exonuclease activity is expressed in two ways when ψ is used as a substrate, which may arise from the presence of two conformations when ψ binds to the enzyme reaction site. The authors then consistently observed the effect of ψ on the residence time of the exonuclease active site in a range of sequences, with the phenomenon being most evident in the 5'-Gψ sequence. This provides an alternative method of sequencing modifications that are not dependent on protein nanopore sensors.
The authors analyzed publicly available RNA sequencing data for SARS-CoV-2 obtained from cell cultures and identified five conserved ψ-sites in the genome. This study extends the understanding of the ψ virus episodic transcriptome and could be used for future interventions and understanding of individual host responses to SARS-CoV-2 virus infection.
BOC Sciences' support for scientific research
PseudoUridine modification holds great promise as a new paradigm for mRNA-based therapeutics. More new insights into the mechanism and function of pseudoUridine modification are needed. BOC Sciences, as the world's largest and most productive supplier, will comply with Good Manufacturing Practices to provide pseudoUridine and abundant pseudoUridine derivatives to customers.
References
- Fleming AM; et al. Nanopore dwell time analysis permits sequencing and conformational assignment of pseudoUridine in SARS-CoV-2. ACS Cent Sci. 2021 Oct 27; 7(10): 1707-1717.
- Begik; et al. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat Biotechnol. 2021; 39, 1278–1291.