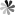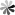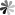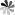Introduction to PseudoUridine
PseudoUridine modifications are the most abundant modifications on intracellular RNA. PseudoUridine is derived from uridine by a base isomerization reaction in which the nucleobase is rotated 180° around the N3-C6 axis, resulting in a change in the nucleobase-glycosidic bond (from N1-C1' to C5 -C1' bond), the resulting C-C bond allows the nucleobase to rotate more freely. PseudoUridine modifications are abundant on tRNA, rRNA, srRNA and other non-coding RNAs with high intracellular abundance. In recent years, with the development of sequencing technology, it has been found that pseudoUridine modifications are also abundant in RNA. PseudoUridine can base pair with adenosine like uridine, but pseudoUridine can alter RNA structure by improving base pairing, base stacking, and backbone stability.
Biological Functions of PseudoUridine
The human immune system recognizes unmodified single-stranded mRNA as a marker of viral infection and activates downstream pathways to block mRNA translation. The recognition of mRNA by the human immune system is mainly based on the recognition of the uridine structure in the mRNA sequence, leading to the development of a technology for replacing uridine with pseudoUridine. This technique not only greatly reduces the immunogenicity of mRNA, but also the immunogenicity of RNA decreases as the proportion of pseudoUridine introduced increases. When pseudoUridine completely replaces uridine, it also improves the stability of mRNA and enhances its translation ability.
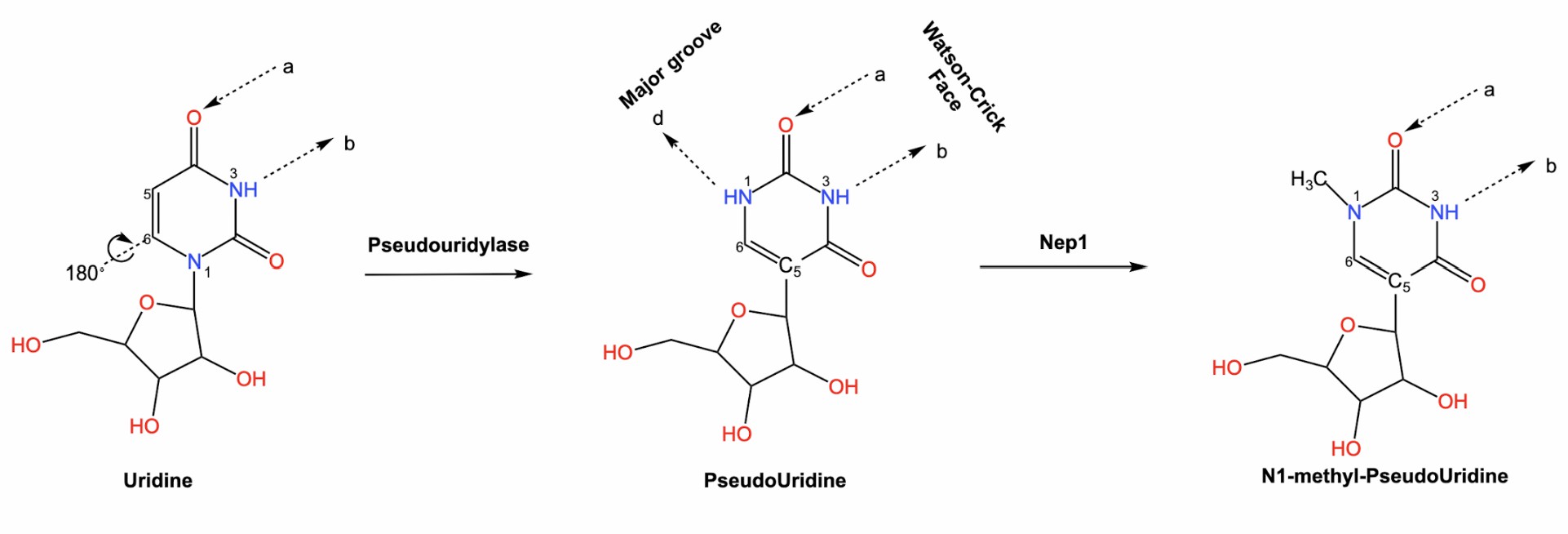 Fig. 1 Structure diagram of pseudoUridine and N1-methyl-pseudoUridine
Fig. 1 Structure diagram of pseudoUridine and N1-methyl-pseudoUridine
| Name | CAS | Symbol |
| pseudoUridine | 1445-07-4 | Ψ |
| N1-methyl-pseudoUridine | 13860-38-3 | m1Ψ |
snRNA
Small nuclear RNA (snRNA) is the main component of the RNA spliceosome during post-transcriptional processing in eukaryotes. Five types of snRNAs have been identified, which are approximately 100-215 nucleotides in length in mammals. snRNA existed in the nucleus, and together with about 40 kinds of intranuclear proteins form the RNA spliceosome, which plays an important role in the post-transcriptional processing of RNA.
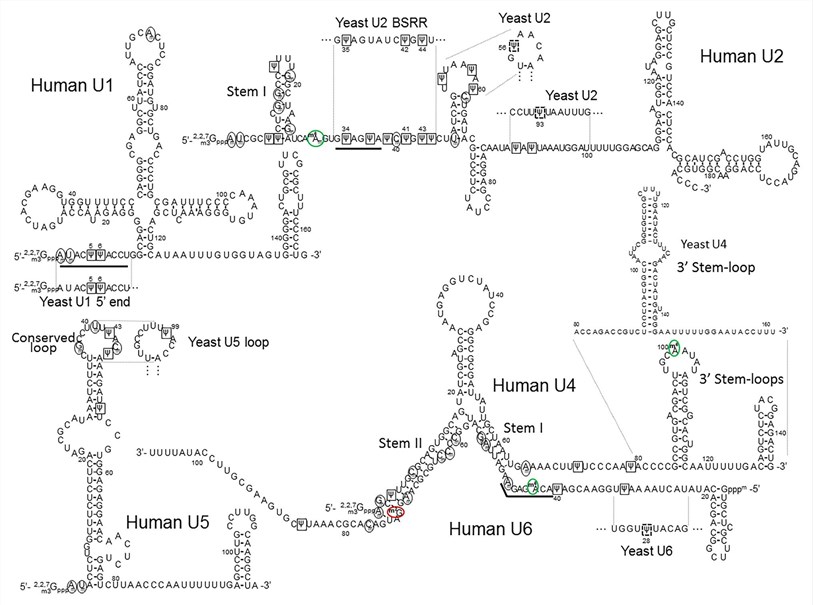 Fig. 2 The primary sequences and the secondary structures of minor class snRNAs (Morais P, 2021)
Fig. 2 The primary sequences and the secondary structures of minor class snRNAs (Morais P, 2021)
PseudoUridine Modifications in snRNA
During pre-mRNA splicing, spliceosomes formed by snRNA (U1, U2, U3, U4, U5, U6) and a series of proteins play an important role in intron splicing. PseudoUridine modifications occur basically in all snRNAs (U2 contains the most pseudoUridine modification sites), and these modifications are highly conserved across species.
Similar to the distribution of pseudoUridines in rRNA, pseudoUridines in snRNA are mainly distributed in functionally important regions. For example, pseudoUridine modifications in U1 snRNA are distributed in the 5' terminal region, which is important for the recognition of the 5' cleavage site during early cleavage. The distribution of pseudoUridine modification in snRNA also illustrates its important function for splicing RNA, and the current study shows that pseudoUridine modification on snRNA acts mainly through RNA-RNA interactions during the splicing process.
The role of pseudoUridine modifications in snRNA can be studied by three systems.
- In vitro systems
- Xenopus laevis microinjection system
- Yeast system
In the in vitro system, the synthesized snRNA was co-incubated with HeLa cell extracts to undergo pseudoUridine modification. 5-fluorouridine is an inhibitor of pseudoUridine formation, and when 5-fluorouridine replaces uridine in RNA, pseudoUridine modification cannot occur at this site. When the in vitro synthesized snRNA incorporates 5-fluorouridine, since normal pseudoUridine modification cannot occur, the snRNA without pseudoUridine modification will not be able to form normal snRNP. Using the Xenopus laevis microinjection system, in vitro synthesized U2 snRNA without pseudoUridine modification was injected into U2-deficient Xenopus eggs and found to affect pre-mRNA splicing. Therefore, pseudoUridine modification in U2 snRNA plays an important role in snRNP formation, spliceosome assembly, and pre-mRNA splicing.
Reference
- Morais P; et al. Spliceosomal snRNA Epitranscriptomics. Front Genet. 2021 Mar 2; 12: 652129.