With the success of BNT162b2 and mRNA-1273 vaccines, mRNA-LNP has taken center stage in vaccine therapy. The mRNA-LNP vaccine is an efficient modular vaccine platform that enables the rapid manufacturing of novel vaccines with optimized designs built on therapeutic targets. Currently, mRNA-LNPs are evaluated based on the immunogenicity of intracellular RNA sensors that recognize exogenous mRNAs, the ability to express encoded mRNAs, as well as the stability and toxic effects throughout LNP preparations. All processes are checked according to treatment goals and route of administration.
mRNA-LNP designing elements
- mRNA sequence design and nucleotide modification selection
- Optimization of LNP formulations to encapsulate and deliver mRNA
- Long-term storage
Key considerations for mRNA sequence and modification selection
- Expression of encoding mRNA
- Immunogenicity induced by intracellular RNA sensors (recognition of mRNA as foreign entities)
- mRNA stability
These factors are controlled by designing mRNAs with modified nucleotides, sequence modifications, and mRNA capping (Figure 1).
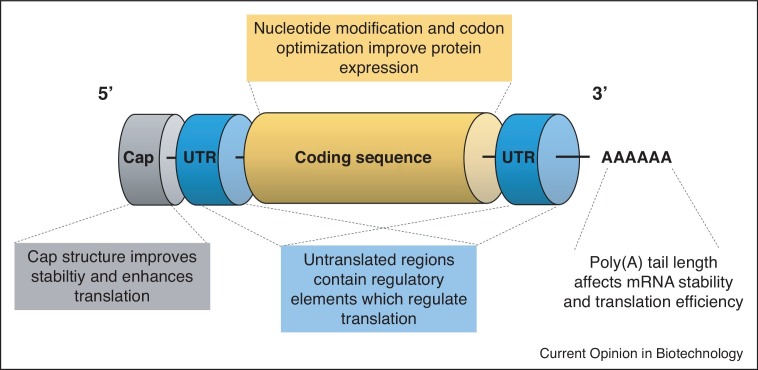 Fig. 1 Optimizing mRNA sequence and modifications for mRNA-LNP vaccines (Kon E, 2022)
Fig. 1 Optimizing mRNA sequence and modifications for mRNA-LNP vaccines (Kon E, 2022)
Nucleotide modification
Based on research studies and existing experiences, including the Pfizer/BioNTech's BNT162b2, Moderna's mRNA-1273, and SARS-CoV-2 mRNA vaccines, nucleotide modification is now considered to be the most important breakthrough in mRNA therapy. Mechanistically, innate immunity is activated after the recognition of non-modified mRNA molecules with cellular RNA sensors. While activation of innate immune pathways with adjuvants may be beneficial in the context of vaccination, these innate immune responses dramatically reduce mRNA translation and lead to a potential detriment in mRNA therapy. In 2005, a pioneering study by Kariko and colleagues demonstrated that the immunogenicity of mRNA could be significantly reduced by the incorporation of naturally occurring-chemically modified nucleosides, such as pseudouridine (ψ), 5-methylcytidine (m5C), and thiouridine (s2U). Further studies have demonstrated the enhancement of RNA stability with the incorporation of modified nucleosides, including protein modification, in which N1-methylpseudouridine modification is employed in both mRNA-1273 and BNT162b2. In contrast, CureVac's sequence-optimized vaccine candidate, CVnCoV, consists entirely of unmodified nucleosides. This unmodified mRNA sequence engineering approach provides a robust and balanced immune response. Further preliminary Phase 2b/3 data indicates that this candidate, CVnCoV, was significantly less effective than the other two leading mRNA vaccines with Pfizer/BioNTech and Moderna. While the inclusion of unmodified nucleosides can lead to inferior efficacy of CureVac vaccine candidates, other dissimilarities between vaccine candidates should also be considered, such as differences in non-coding elements and storage conditions (Figure 2). Currently, both FDA-licensed mRNA vaccines with Pfizer/BioNTech and Moderna contain modified nucleotides.
| BNT162b2 (Pfizer/BioNTech) | mRNA-1273 (Moderna) | CVnCoV (CureVac) | |
| Approval Status | FDA Approved August 23, 2021 | Emergency Use Authorization (EUA) December 18, 2020 | Phase 2b/3 (NCT04652102) |
| Submitted for full Approval August 25, 2021 | |||
| Lipid Formulation (% Molar ratio of Ionizable: Cholesterol: Neutral lipid: | 46.3: 42.7: 9.4: 1.6 | 50: 38.5: 10: 1.5 | 50: 38.5: 10: 1.5 |
| N: P Molar Ratio | 6 | ||
| Cap | Cap-1 | ||
| 5’-UTR | 5’-untranslated region derived from human alpha-globin RNA with an optimized Kozak sequence | Undisclosed | 5’ UTR: Artifacts from restriction and transcription site, plus Kozak sequence |
| S protein antigen | Codon-optimized sequence encoding full-length SARS-CoV-2 spike (S) glycoprotein containing mutations K986P and V987P | Codon-optimized sequence encoding full-length SARS-CoV-2 spike (S) glycoprotein containing mutations K986P and V987P | Codon-optimized and engineered sequence encoding full-length SARS-CoV-2 spike (S) glycoprotein containing mutations K986P and V987P |
| Modified nucleotides | N1-methyl-pseudouridine | N1-methyl-pseudouridine | Unmodified nucleotides |
| 3’-UTR | 3’ UTR comprising two sequence elements derived from the amino-terminal enhancer of split (AES) mRNA and the mitochondrial encoded 12S ribosomal RNA | Undisclosed | 3’ UTR comprising human alpha-globin 3’ UTR sequence element |
| Poly (A) Tail | A 110-nucleotide poly (A)-tail consisting of a stretch of 30 adenosine residues, followed by a 10-nucleotide linker sequence and another 70 adenosine residues | Undisclosed | 64 Poly (A) tail |
| Storage | -60°C to -80°C for six months, -15°C to -25°C for two weeks, 2°C - 8°C for 5 days | -15°C to -20°C for six months, 2°C - 8°C for 30 days | 2°C - 8°C for three months |
| Buffer | PBS | Tris | Undisclosed |
| Cryoprotectant | Sucrose | Sucrose | Undisclosed |
Table 1 Comparison of the mRNA and LNP formulation components of three SARS-CoV-2 mRNA-LNP vaccines (Kon E, 2022)
References
- Kon E; et al. Principles for designing an optimal mRNA lipid nanoparticle vaccine. Curr Opin Biotechnol. 2022 Feb; 73: 329-336.
- Karikó K; et al. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005 Aug; 23(2): 165-75.