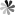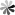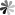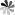Natural RNAs are comprised of a large number of modified nucleosides such as the C-glycoside pseudouridine(Ψ). Pseudouridine is the most common modified nucleotide and is universally conserved in several RNAs at regions of biological importance. 15N labels allow overlapping 1H NMR resonances to be resolved and can serve as local probes of hydrogen bonding or protonation because they display a wide range of chemical shifts that are sensitive to small changes in the local environment. The site-specific incorporation of 15N labels is important for the structural analysis of RNA or the study of ligand interactions with nucleic acids.
However, the use of biological methods still requires the chemical synthesis of 15N-labeled bases before they are added as 15N sources to culture media for bacterial strains capable of incorporating 15N into the desired biomolecule, thereby extracting the interest biomolecule enriched with 15N. This biosynthetic method works well for the isolation of total RNAs, whereas the isolation of smaller RNA fragments is generally tedious and the quantity obtained may not be sufficient for NMR structural analysis. Nucleotides can be efficiently coupled using solid-phase chemistry to yield short polynucleotides in milligram quantities. The four standard RNA nucleosides have been labeled with 15N through a wide variety of synthetic techniques. PseudoUridine, as a modified nucleoside, also can be enrich with 15N site-specifically by chemical synthesis method.
Synthesis of 15N-labeled pseudoUridine
Tetraisopropyl disiloxane (TIPDS) can be used to protect both the 2’- and 5’- hydroxyl groups on the ribose. The protective groups can be employed in the synthesis of 15N-enriched pseudoUridine derivatives. The chemical synthesis route from 3′,5′-O-TIPDS-pseudoUridine to [3-15N-methyl]-3′,5′-O-TIPDS-pseudoUridine and [3-15N]-3′,5′-O-TIPDS-pseudoUridine, which serve as precursors to the corresponding phosphoramidites, has been reported. The initia lcompound was first acetylated with acetic anhydride in basic condition and then the N1 position was protected with pivaloyloxymethyl (POM). A mild nitrating reagent, nitronium trifluoroacetate (NO2OCOCF3), was then implemented for the generation of the N-nitro nucleosides. Then, the ammoniolysis reaction was carried out with 15NH4Cl and K2CO3 to yield compound labeled with 15N. Methylation at 3-15N can be accomplished by using N,N-dimethylformamide dimethyl acetal. The advantage of using 2′-OAc and N1-POM protective groups is that they can be removed in one step by treating them with 2.0 M NH3 in MeOH and heating to 50 °C.
Application
PseudoUridine 1915 of E. coli 23S rRNA is naturally methylated at the N3 position. These labeled nucleosides are important for probing the biological roles of pseudoUridine and 3-methylpseudoUridine. The 15N-labeled pseudoUridine will have other practical applications. The incorporation of pseudoUridine into oligonucleotides will help determine the RNA structure by NMR spectroscopy, to test the pKa of the NH amines within the context of short RNAs, to exam hydrogen-bonding interactions in RNA duplexes, and to screen for small-molecule-RNA or RNA-RNA interactions.
Reference
- Desaulniers, J. P., Ksebati, B., & Chow, C. S. (2003). Synthesis of 15N-enriched pseudouridine derivatives. Organic Letters, 5(22), 4093-4096.
-9-hydroxy-2,2,4,4-tetraisopropyltetrahydro-6h-furo[3,2-f][1,3,5,2,4]trioxadisilocin-8-yl)-1,3-dimethylpyrimidine-2,4(1h,3h)-dione-3-15n.gif)
-9-hydroxy-2,2,4,4-tetraisopropyltetrahydro-6h-furo[3,2-f][1,3,5,2,4]trioxadisilocin-8-yl)-1-methylpyrimidine-2,4(1h,3h)-dione-3-15n.gif)