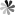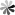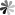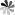Post-transcriptional modifications are a component of naturally occurring functional RNAs. PseudoUridine (Ψ) that known as the fifth RNA residue is the most common modification in RNA. PseudoUridine is commonly observed in ribosomal RNA (rRNA), and it is prevalent in transfer RNA (tRNA), small nuclear RNA (snRNA) and other noncoding RNAs (ncRNAs). In addition, the discovery of the presence of PseudoUridine in yeast and human messenger RNAs (mRNAs) encourages people to explore the probable effects of PseudoUridine in mRNA. Recently, the importance of PseudoUridine in epigenetic modulation of mRNA function is becoming increasingly apparent. The functions of pseudoUridine and its derivatives, whether naturally occurring or chemically synthesized, remain to be discovered in many of the cases.
Naturally occurring pseudoUridine derivatives in RNA modification
The role of pseudoUridine structure has been well investigated both experimentally and theoretically. Interestingly, RNAs also contain modifications, which are derivatives of pseudoUridine. There are four pseudoUridine derivatives discovered in natural RNAs modification, including 1-methylpseudoUridine (m1Ψ), 3-methylpseudoUridine (m3Ψ), 2′-O-methylpseudoUridine (Ψm), and a hypermodified residue 1-methyl-3-(3-amino-3-carboxypropyl) pseudoUridine (m1acp3Ψ). These modified pseudoUridines are produced by derivatization of the hydroxy group on ribose or amino in the uracil base of pseudoUridine.
Function of modified pseudoUridine
The m1Ψ was first observed in the tRNAs of archaebacteria in which were found to occur in the TΨC loop in place of thymidine. Studies have been indicated that m1Ψ modified mRNA has a good performance to enhance protein expression, and m1Ψ used for the modification of vitro-transcribed mRNA which applicated in RNA therapy exhibited non cytotoxicity at different concentrations. The m3Ψ was first identified in a 23S rRNA, a region that is a component of the site of protein synthesis in the ribosome. Selective introduction of a methyl group at the N3 position of pseudoUridine at site 1915 of 23S RNA results in a slight enhancement in the thermodynamic stability of the RNA compared to pseudoUridine. The 2′-O-methylated derivative of Ψ has been observed in tRNAs of archaea and eukaryotes, and in snRNAs in eukaryotes. In general, the 2′-O-methylation of the ribose stabilizes the C3′- endo conformation and makes the base pairing more thermodynamically stable in RNA. The m1acp3Ψ was first isolated from the RNA in an established line of cells of the Chinese hamster in 1974. Recently, it has been found that deletion of m1acp3Ψ rRNA modification is a major feature of cancer.
Chemically synthesized modified pseudoUridine
In addition, the derivatization of pseudoUridine through chemical synthesis can also be used for the modification of RNA to improve its function. These chemically synthesized modified pseudoUridines were reported in several literatures. For example, 4-thiopseudoUridine containing thiocarbonyl group was found to stabilize the triplex formation of triplex-forming oligodeoxynucleotides (TFOs), which is means that a particular DNA or RNA oligonucleotide bind to the high pyrimidine region of DNA via the formation of hydrogen bonds. ThiopseudoUridine can be used to form highly stable parallel triplex structures likely through the strong stacking interaction of the thiocarbonyl group with the 5’-upstream or 3’-downstream bases of TFOs.
In conclusion, despite intensive investigation of the structural and biochemical effects of pseudoUridine in various systems, the biological roles of most pseudoUridine and its derivatives remain unknown. The challenging questions relate to PseudoUridine function in the context of cellular RNA need to be explored and solved further.
pseudouridine.gif)

pseudouridine.gif)










-n1-methylpseudouridine.gif)
pseudouridine.gif)
-3,4-dihydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl)-3-(4-hydroxybutyl)-1-methylpyrimidine-2,4(1h,3h)-dione.gif)
propyl pseudouridine.gif)